TRANSFAC download
Software demo: Coffee break with TRANSFAC
May 28th, 10 AM CEST
FREE online seminar
Seminar Description
Download TRANSFAC and do whatever you like
Introduction to the TRANSFAC flat file download release
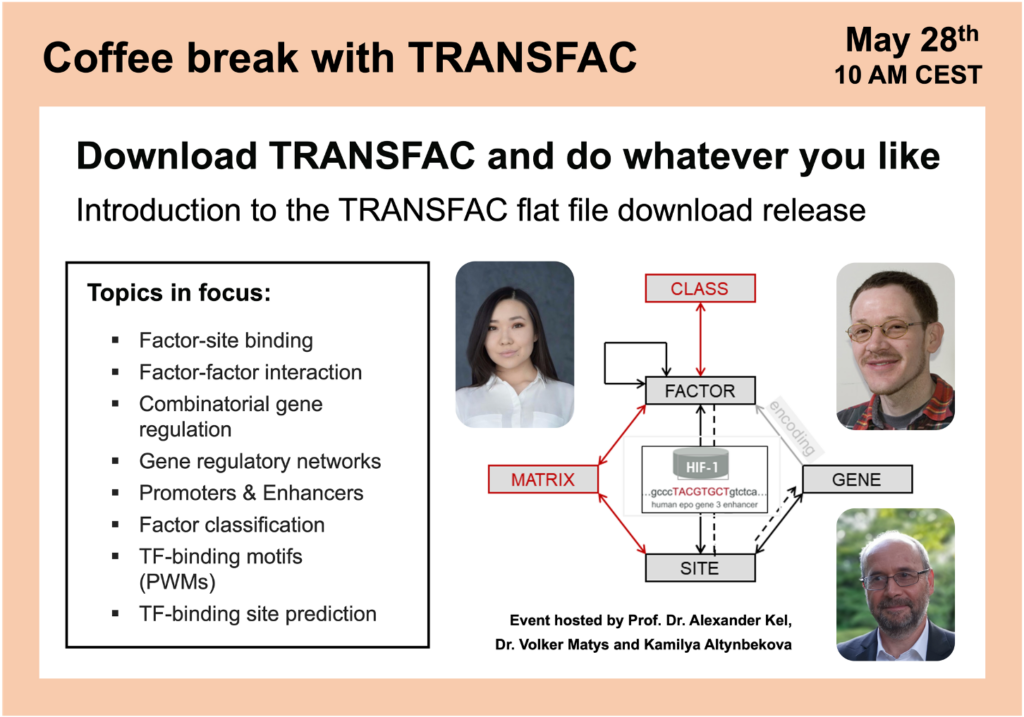
TRANSFAC® (including TRANSCompel® and TANSPro™) contains eukaryotic transcription factors (and miRNAs), their experimentally determined genomic binding sites and consensus DNA-binding motifs (PWMs), as well as data on combinatorial gene regulation and factor-factor interaction.
Promoters, enhancers and silencers annotated with transcription factor ChIP-Seq, DNase hypersensitivity and histone methylated intervals from the ENCODE project and from other sources complement the manually curated binding site data.
Based on the positional weight matrices (PWMs) transcription factor binding sites can be predicted in regulatory regions (with Match™).
In the TRANSFAC® flat file download, the Match™ tool can be used in the command line or the PWMs can be used with third-party tools or with tools of the user.
This webseminar will give a brief overview of TRANSFAC® with the focus on the flat file download release.
The following main topics will be in the focus of this webseminar:
What you will learn in this webseminar
Seminar Schedule
The session will start on May 28 with a lecture and software demo from Dr. Volker Matys and Kamilya Altynbekova (approx. 1 hour) and will be continued with a Q&A session addressing the questions coming from the attendees of the online event (up to 30 minutes).
What You Get
The Lecturers

Prof. Dr. Alexander Kel

Kamilya Altynbekova

Dr. Volker Matys
Prerequisites
Basic knowledge of biochemistry and molecular biology. Being familiar with the use of Java-based or other command line tools. Data extraction from the flat files requires programming skills (or SQL).


